- Home
- Products
- Pathway
- Support
- Contact Us

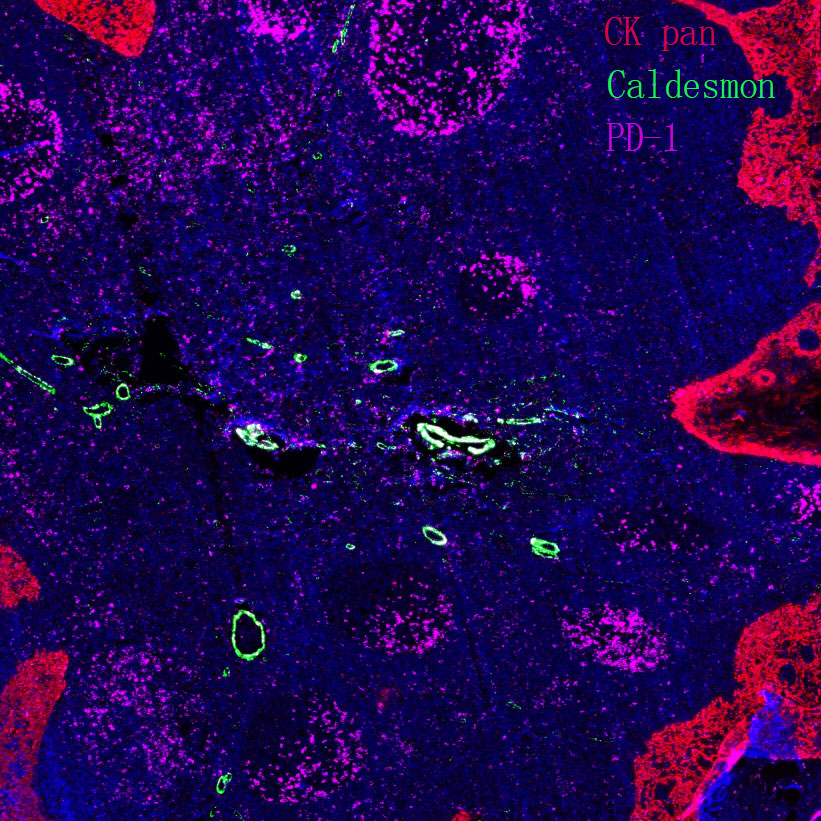
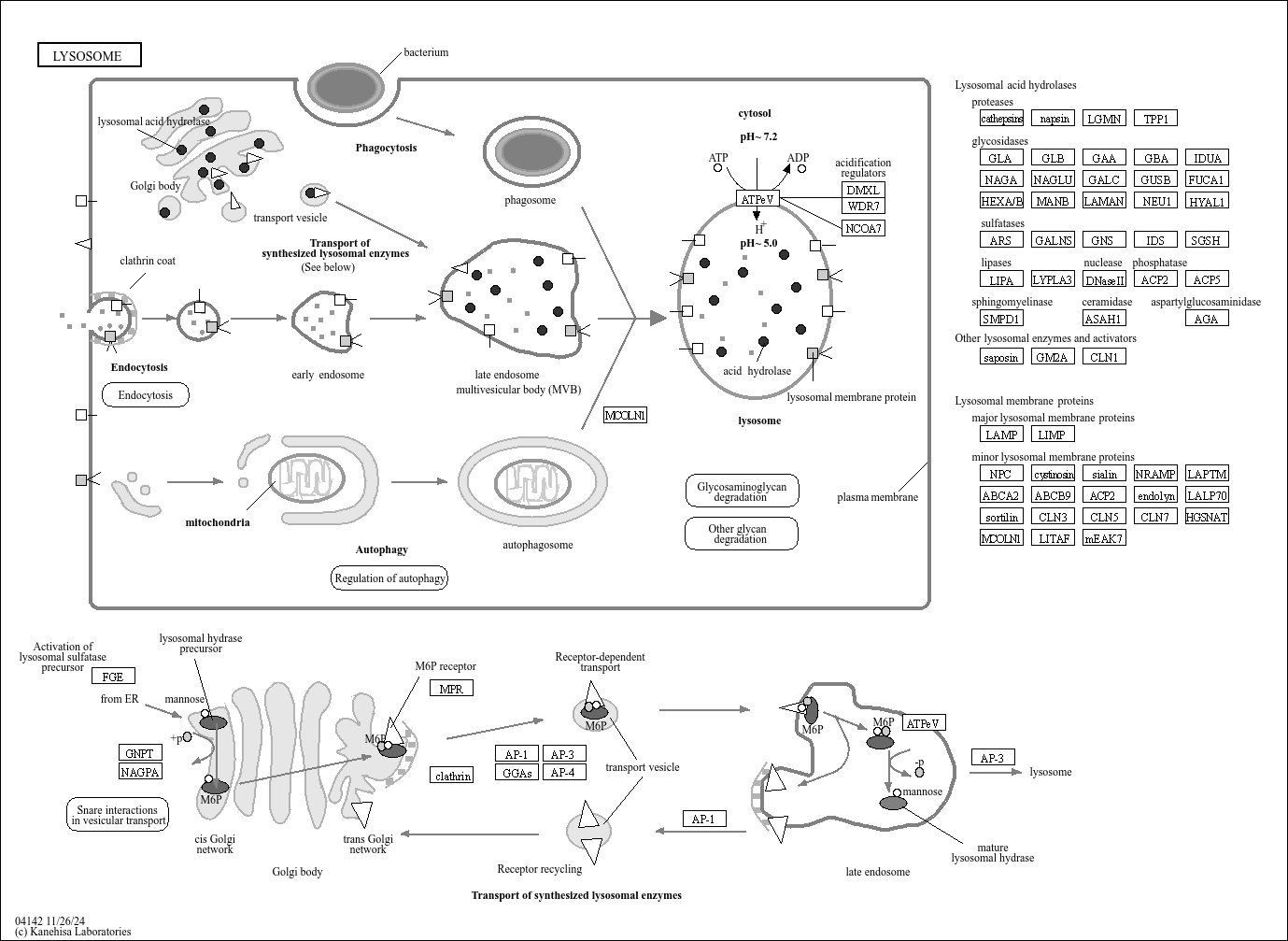
Lysosome
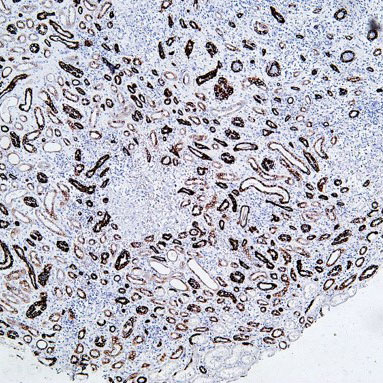
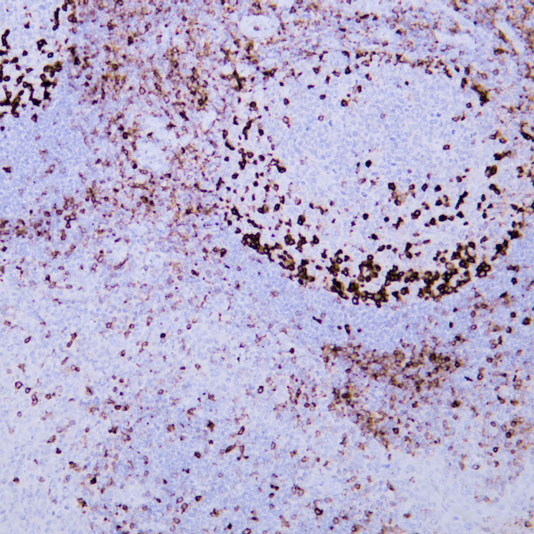
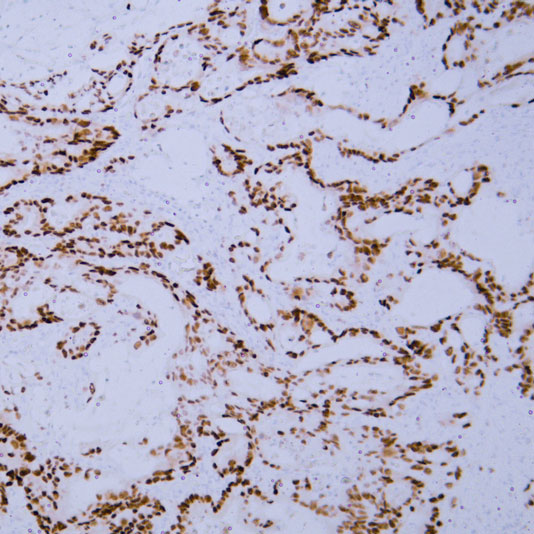
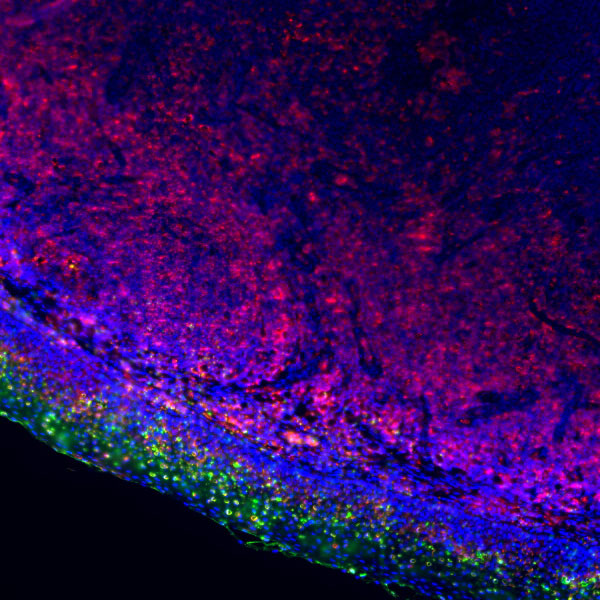
Core of basic research: Clarifies the structural and functional mechanisms of lysosomes as "cellular digestive workshops," the core organelles for intracellular substance degradation, nutrient recycling, and signal regulation. Lysosomes are membrane-bound organelles containing various acid hydrolases (proteases, nucleases, glycosidases, etc.); V-ATPase on the membrane actively transports protons to maintain an acidic internal environment (pH 4.5-5.0), ensuring hydrolase activity. Core functions include: Fusing with autophagosomes to form autolysosomes for degrading damaged organelles or abnormal proteins; degrading extracellularly internalized pathogens or nutrients via the endocytic pathway; releasing hydrolases through lysosomal membrane permeabilization (LMP) to regulate apoptosis or necroptosis; acting as a metabolic signal sensing platform (e.g., mTORC1 complex localized on the lysosomal membrane to sense amino acid concentration) to regulate cell growth and autophagic balance. Research focuses include the synthesis and sorting mechanisms of lysosomal enzymes, V-ATPase-mediated acidification regulation, lysosomal membrane stability control, interactions between lysosomes and other organelles (mitochondria, endoplasmic reticulum), and the association of pathway abnormalities with lysosomal storage diseases (substrate accumulation due to enzyme deficiency) and neurodegenerative diseases (impaired abnormal protein degradation).
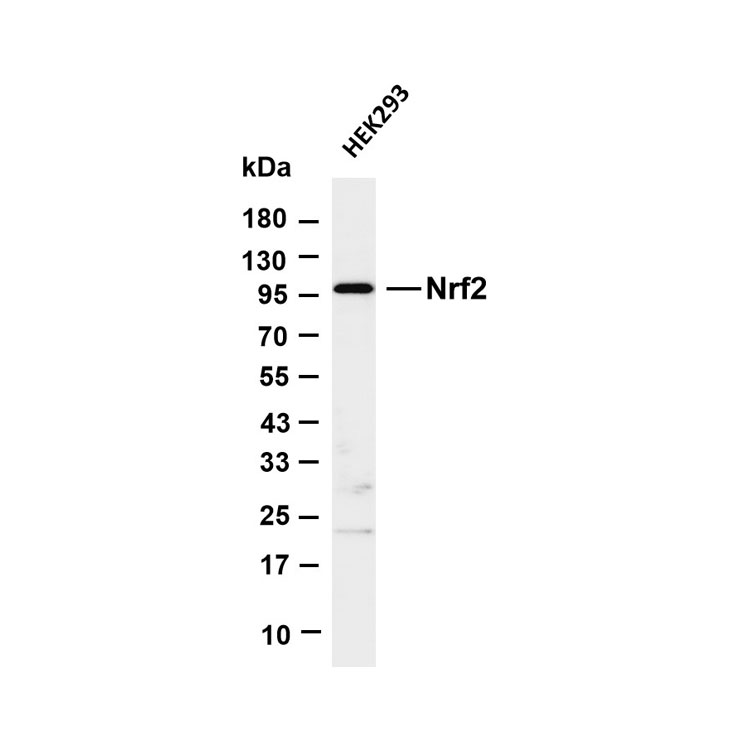
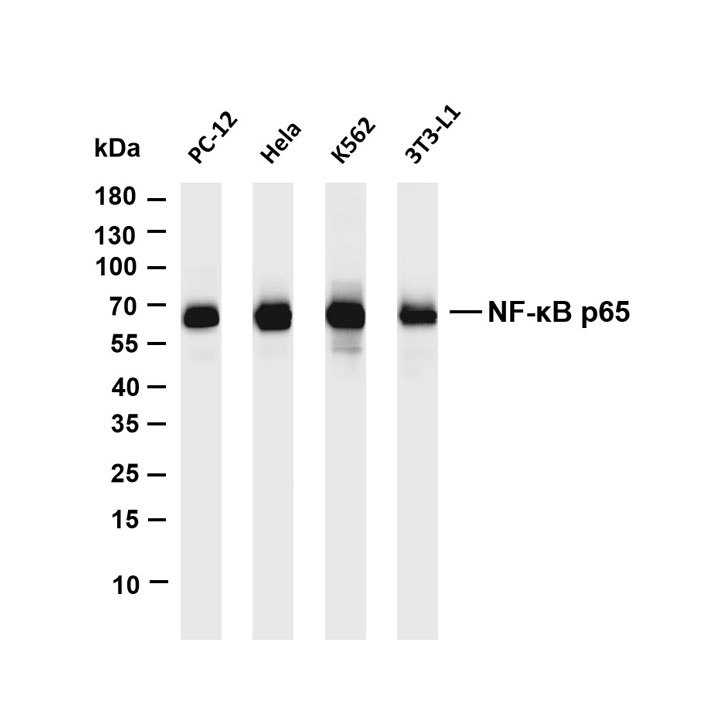
Core key proteins: Acid hydrolases (Cathepsin B/D/L, β-glucocerebrosidase, sphingomyelinase), V-ATPase (lysosomal membrane proton pump maintaining acidic environment), lysosomal membrane proteins (LAMP1/LAMP2, CD63), mTORC1 complex (localized on lysosomal membrane, regulating metabolism), autophagy-related proteins (LC3, p62, mediating autophagosome-lysosome fusion), Rab GTPases (Rab7/Rab5, regulating lysosomal transport and fusion), cathepsin inhibitors (cystatins, regulating hydrolase activity), NPC1/NPC2 (lysosomal cholesterol transporters).
Core key proteins: Acid hydrolases (Cathepsin B/D/L, β-glucocerebrosidase, sphingomyelinase), V-ATPase (lysosomal membrane proton pump maintaining acidic environment), lysosomal membrane proteins (LAMP1/LAMP2, CD63), mTORC1 complex (localized on lysosomal membrane, regulating metabolism), autophagy-related proteins (LC3, p62, mediating autophagosome-lysosome fusion), Rab GTPases (Rab7/Rab5, regulating lysosomal transport and fusion), cathepsin inhibitors (cystatins, regulating hydrolase activity), NPC1/NPC2 (lysosomal cholesterol transporters).
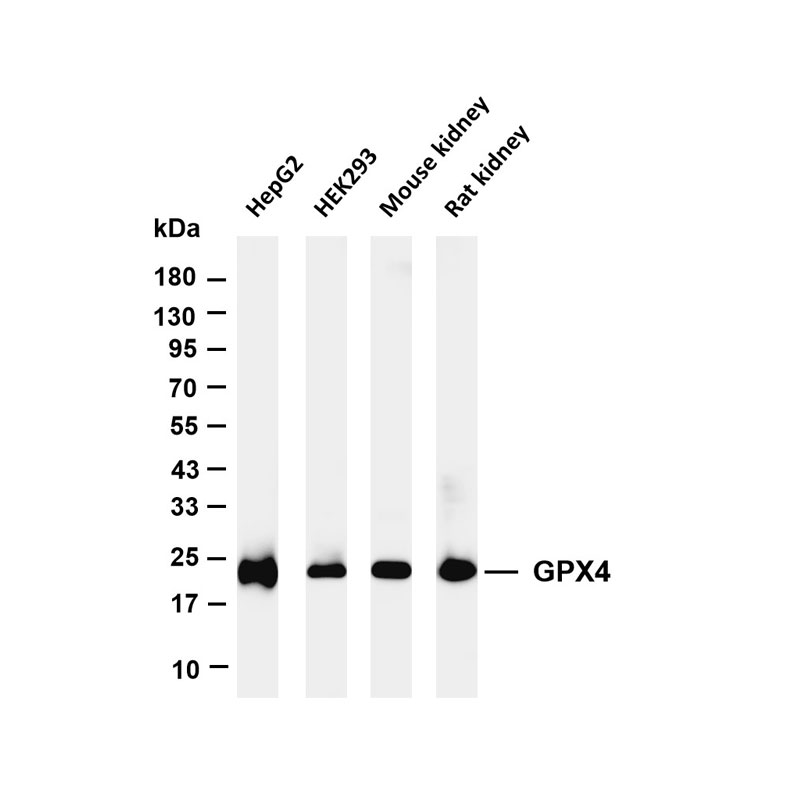
Product list
-
{{item.title}}{{item.react}}{{item.applicat}}
Product list
Product name
Reactivity
Application
Related Resource Links
Related Promotional Journal Downloads

Explore Our Recommended Popular Products
More products
30,000+ high- quality products available online
Primary Antibodies, Secondary Antibodies, mIHC Kits, ELISA Kits, Proteins, Molecular Biology Products,Cell Lines,Reagents ...
Contact Us