- Home
- Products
- Pathway
- Support
- Contact Us

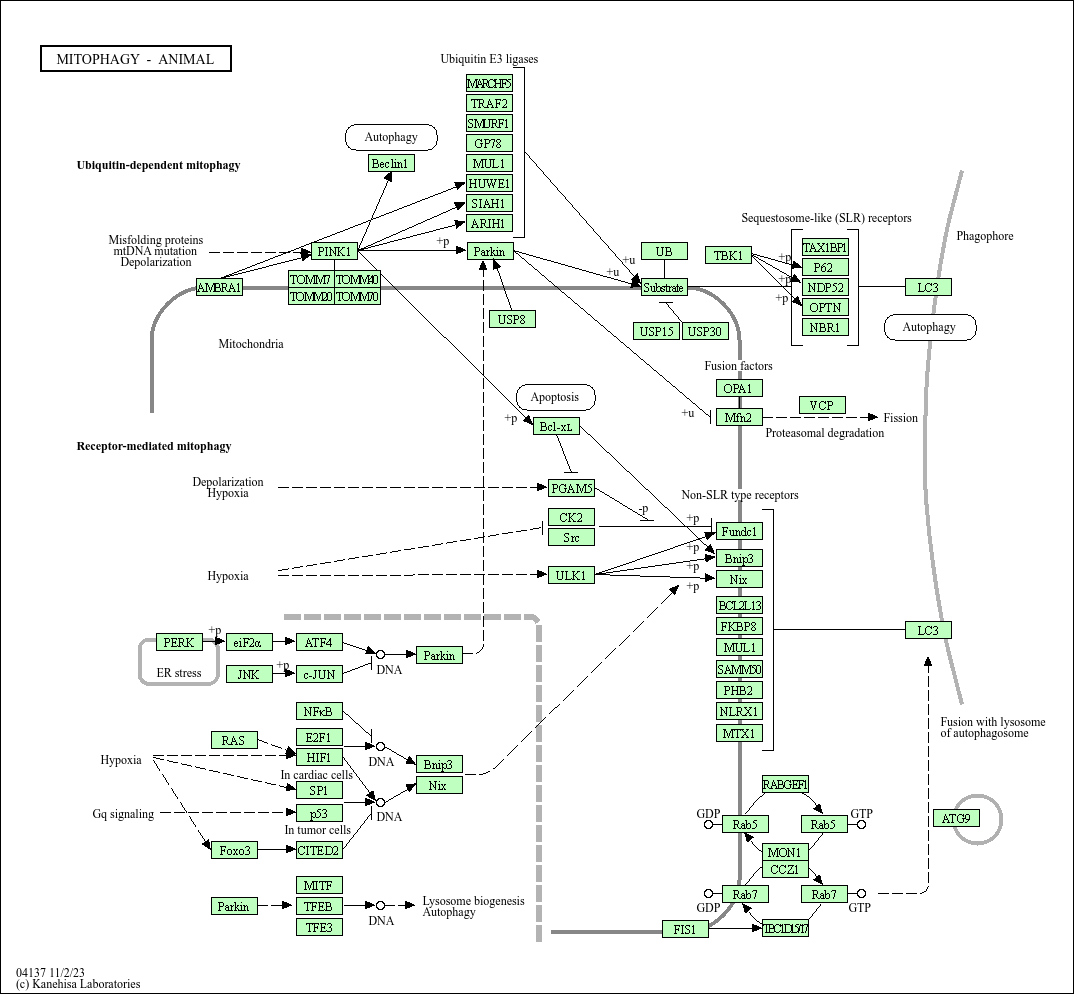
Mitophagy - animal
Core of basic research: Deciphers the molecular mechanism by which animal cells selectively recognize and degrade damaged/excessive mitochondria, critical for maintaining mitochondrial homeostasis and avoiding oxidative stress damage. Core regulatory pathways include ubiquitin-dependent and receptor-dependent types: In the ubiquitin-dependent pathway, mitochondrial damage induces ROS release or reduced membrane potential, triggering the recruitment of Parkin (E3 ubiquitin ligase) to the mitochondrial membrane, which ubiquitinates mitochondrial proteins (e.g., TOM20, VDAC1), further recruiting autophagic receptors such as p62/SQSTM1 to mediate autophagosome encapsulation of mitochondria. In the receptor-dependent pathway, mitochondrial membrane receptors (e.g., BNIP3, NIX/BNIP3L) directly bind the autophagy-related protein LC3, initiating mitochondrial encapsulation without ubiquitination. Eventually, mitochondria-encapsulating autophagosomes fuse with lysosomes to degrade mitochondria and recycle nutrients. Research focuses include mitochondrial damage sensing mechanisms, Parkin recruitment and ubiquitination regulation, selective recognition mechanisms of autophagic receptors, cross-talk between mitophagy and apoptosis, and the association of pathway abnormalities with Parkinson’s disease (Parkin mutations) and myocardial ischemia-reperfusion injury.
Core key proteins: Parkin (E3 ubiquitin ligase), PINK1 (serine/threonine kinase sensing mitochondrial damage and recruiting Parkin), autophagic receptors (p62/SQSTM1, NDP52, OPTN), mitochondrial membrane receptors (BNIP3, NIX/BNIP3L, FUNDC1), LC3 (autophagosome membrane protein binding to receptors), Atg family proteins (Atg5, Atg7, Atg12, participating in autophagosome formation), ubiquitin (marking damaged mitochondria), mitochondrial membrane proteins (TOM20, VDAC1, ubiquitination substrates), Rab7 (regulating autophagosome-lysosome fusion).
Core key proteins: Parkin (E3 ubiquitin ligase), PINK1 (serine/threonine kinase sensing mitochondrial damage and recruiting Parkin), autophagic receptors (p62/SQSTM1, NDP52, OPTN), mitochondrial membrane receptors (BNIP3, NIX/BNIP3L, FUNDC1), LC3 (autophagosome membrane protein binding to receptors), Atg family proteins (Atg5, Atg7, Atg12, participating in autophagosome formation), ubiquitin (marking damaged mitochondria), mitochondrial membrane proteins (TOM20, VDAC1, ubiquitination substrates), Rab7 (regulating autophagosome-lysosome fusion).
Product list
-
{{item.title}}{{item.react}}{{item.applicat}}
Product list
Product name
Reactivity
Application
Related Resource Links
Related Promotional Journal Downloads

Explore Our Recommended Popular Products
More products
30,000+ high- quality products available online
Primary Antibodies, Secondary Antibodies, mIHC Kits, ELISA Kits, Proteins, Molecular Biology Products,Cell Lines,Reagents ...
Contact Us