- Home
- Products
- Pathway
- Support
- Contact Us

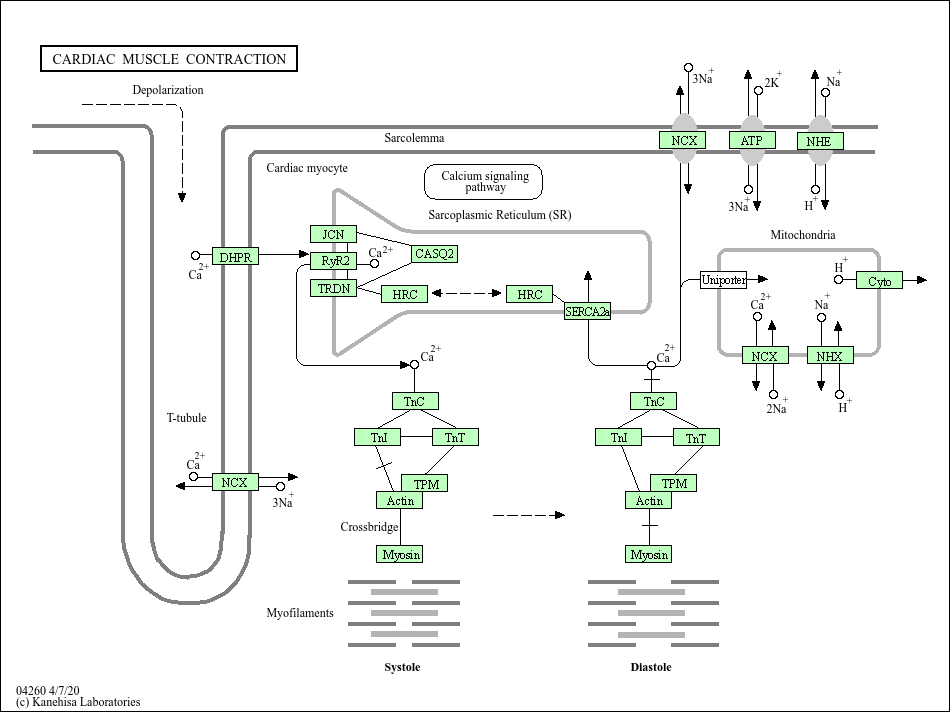
Cardiac muscle contraction
Core of basic research: Clarifies the molecular mechanism of "excitation-contraction coupling" in cardiomyocytes, the process of converting electrical signals into mechanical contraction. When action potentials propagate along cardiomyocyte membranes, LTCC are activated to induce minimal calcium influx. Influxed calcium binds to RyR2 on the SR, triggering massive SR calcium release (CICR) and rapid elevation of cytoplasmic calcium concentration. Calcium binds to troponin C (TnC), inducing conformational changes in the troponin complex to relieve tropomyosin blockage of actin-myosin binding sites. Myosin heads bind to actin, hydrolyze ATP for energy, and undergo conformational changes to drive myofilament sliding, resulting in myocardial contraction. After contraction, cytoplasmic calcium is reuptaken into the SR via SERCA, and residual calcium is extruded via the sodium-calcium exchanger (NCX), reducing cytoplasmic calcium concentration and inducing myocardial relaxation. Research focuses on the regulatory mechanism of CICR, SERCA calcium reuptake efficiency, the molecular motor mechanism of myofilament sliding, load-dependent regulation of myocardial contraction, and mechanisms of contractile dysfunction in heart failure (e.g., calcium cycling disorders, RyR2 leakage).
Core key proteins: Cardiac actin, cardiac myosin, troponin (TnC, TnI, TnT), tropomyosin, L-type calcium channel (LTCC), RyR2 (SR calcium release channel), SERCA (SR calcium ATPase), PLB (phospholamban regulating SERCA activity), CaM (calmodulin), sodium-calcium exchanger (NCX), ATP (energy donor), cardiomyocytes, sarcoplasmic reticulum.
Core key proteins: Cardiac actin, cardiac myosin, troponin (TnC, TnI, TnT), tropomyosin, L-type calcium channel (LTCC), RyR2 (SR calcium release channel), SERCA (SR calcium ATPase), PLB (phospholamban regulating SERCA activity), CaM (calmodulin), sodium-calcium exchanger (NCX), ATP (energy donor), cardiomyocytes, sarcoplasmic reticulum.
Product list
-
{{item.title}}{{item.react}}{{item.applicat}}
Product list
Product name
Reactivity
Application
Related Resource Links
Related Promotional Journal Downloads

Explore Our Recommended Popular Products
More products
30,000+ high- quality products available online
Primary Antibodies, Secondary Antibodies, mIHC Kits, ELISA Kits, Proteins, Molecular Biology Products,Cell Lines,Reagents ...
Contact Us