- Home
- Products
- Pathway
- Support
- Contact Us

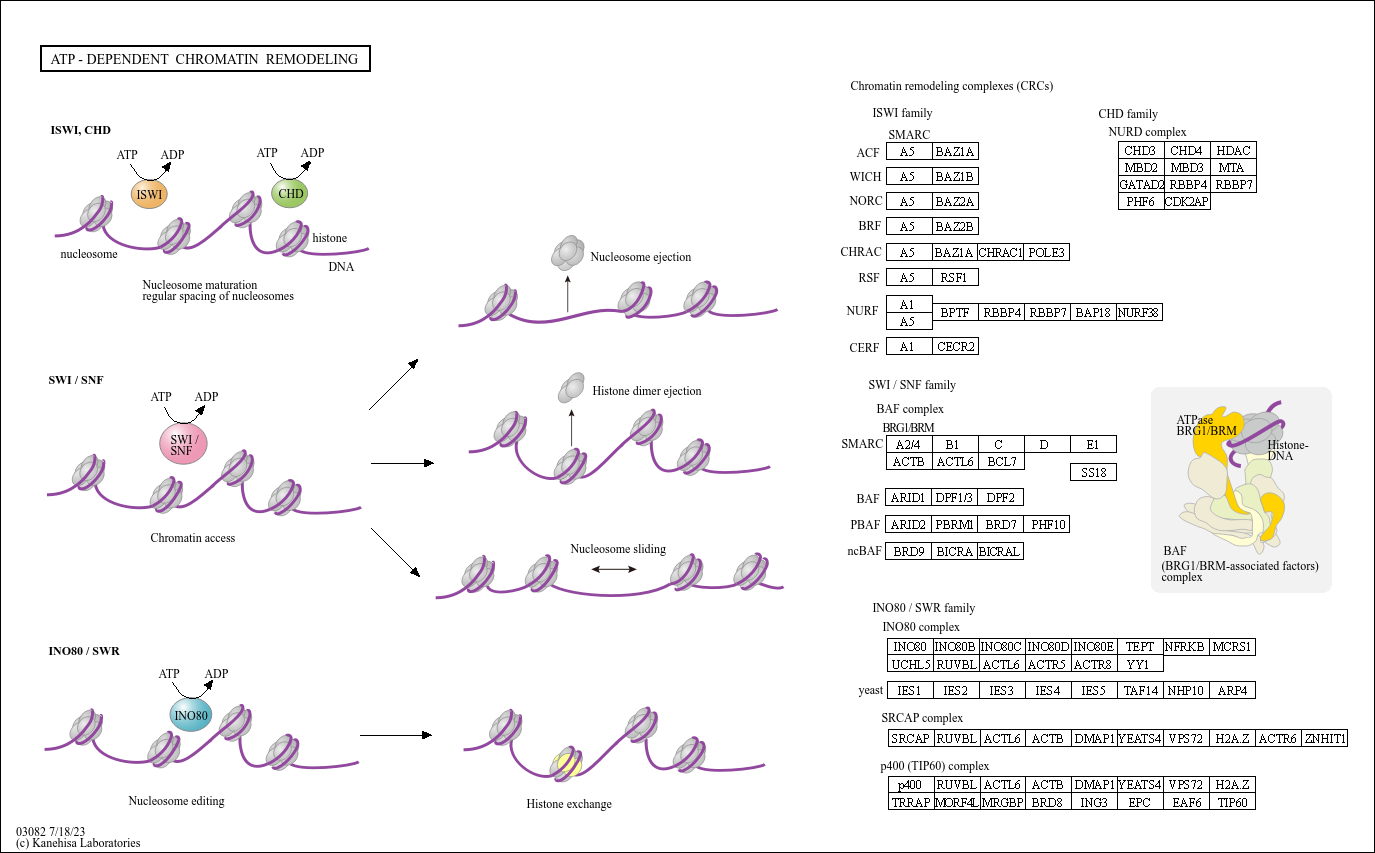
ATP-dependent chromatin remodeling
Core of basic research: Focuses on the molecular mechanisms by which ATP-dependent chromatin remodeling complexes regulate dynamic changes in chromatin structure, a key process for gene transcription regulation, DNA replication, and repair. Remodeling complexes (e.g., SWI/SNF, ISWI, CHD, INO80 families) hydrolyze ATP to provide energy, driving nucleosome sliding, assembly, disassembly, or histone variant exchange, thereby altering chromatin condensation status: decondensing chromatin in silent gene regions (open conformation) to facilitate transcription factor binding and gene activation; or condensing chromatin in active gene regions (closed conformation) to inhibit gene expression. Research focuses include the functional specificity of different remodeling complexes, the synergistic effect between chromatin remodeling and epigenetic modifications (histone acetylation, DNA methylation), and the association of pathway abnormalities with tumors (e.g., SWI/SNF complex mutations) and developmental defects.
Core key proteins: SWI/SNF complex (subunits BRG1, BRM, BAF155, BAF170), ISWI complex (subunits SNF2H, SNF2L), CHD complex (CHD1-CHD9), INO80 complex (subunits INO80, SWR1), histone variants (H2A.Z, H3.3), ATPase subunits (catalyze ATP hydrolysis), transcription factors (bind to remodeled chromatin), DNA polymerase (associated with DNA replication), DNA repair proteins (associated with damage repair).
Core key proteins: SWI/SNF complex (subunits BRG1, BRM, BAF155, BAF170), ISWI complex (subunits SNF2H, SNF2L), CHD complex (CHD1-CHD9), INO80 complex (subunits INO80, SWR1), histone variants (H2A.Z, H3.3), ATPase subunits (catalyze ATP hydrolysis), transcription factors (bind to remodeled chromatin), DNA polymerase (associated with DNA replication), DNA repair proteins (associated with damage repair).
Product list
-
{{item.title}}{{item.react}}{{item.applicat}}
Product list
Product name
Reactivity
Application
Related Resource Links
Related Promotional Journal Downloads

Explore Our Recommended Popular Products
More products
30,000+ high- quality products available online
Primary Antibodies, Secondary Antibodies, mIHC Kits, ELISA Kits, Proteins, Molecular Biology Products,Cell Lines,Reagents ...
Contact Us