- Home
- Products
- Pathway
- Support
- Contact Us

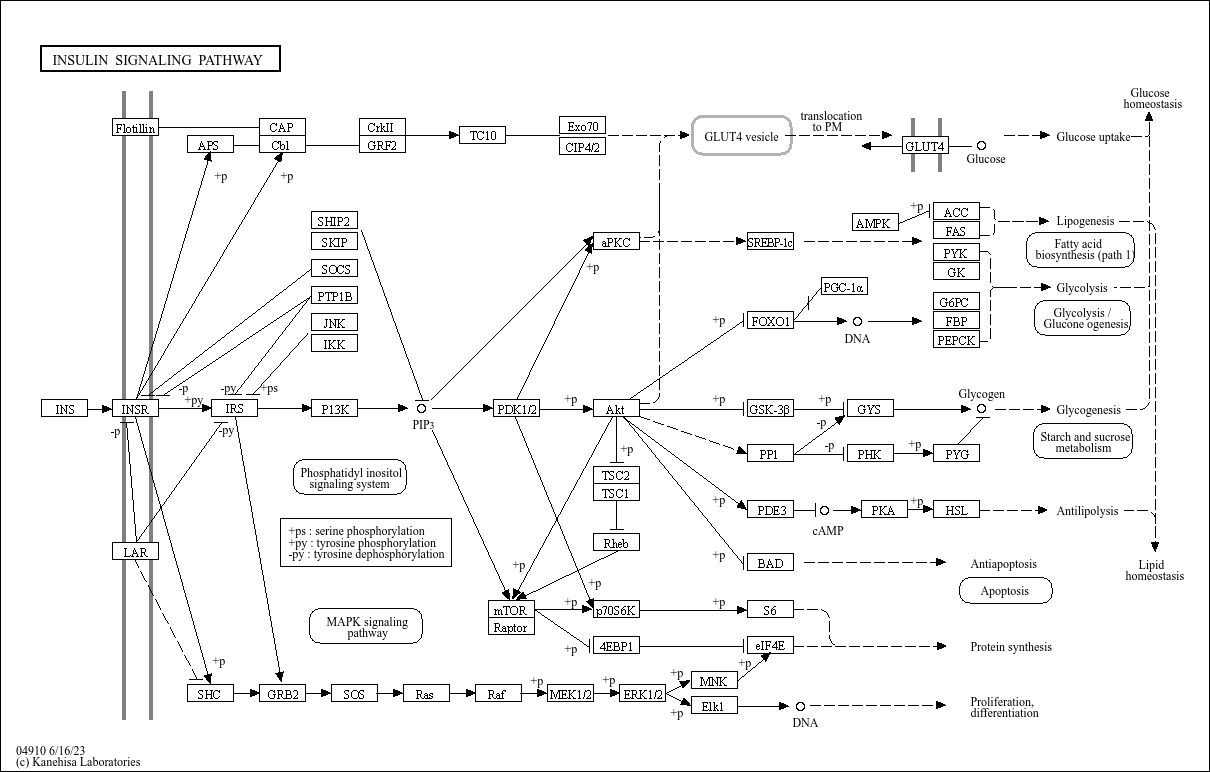
Insulin signaling pathway
Core of basic research: Deciphers the molecular mechanism by which insulin regulates glucose/lipid metabolism and protein synthesis in target cells (liver, muscle, adipocytes), the core pathway maintaining systemic metabolic homeostasis. Insulin binding to the insulin receptor (IR) on target cell membranes induces conformational changes in the α subunit, activating the tyrosine kinase activity of the β subunit, which autophosphorylates and phosphorylates insulin receptor substrates (IRS1/2). Phosphorylated IRS1/2 recruit and activate PI3K, which catalyzes PIP2 to PIP3. PIP3 binds and activates Akt, initiating downstream signals: in adipocytes and muscle cells, Akt promotes GLUT4 vesicle translocation to the membrane, enhancing glucose uptake; in hepatocytes, Akt inhibits gluconeogenic enzymes (PEPCK, G6Pase) and activates glycogen synthase (GS), reducing hepatic glucose output and promoting glycogen storage. Insulin also activates the Ras-MAPK pathway to promote target cell proliferation and differentiation. Research focuses on the phosphorylation regulation of IRS proteins, negative feedback mechanisms (PTEN hydrolyzing PIP3; SOCS3 inhibiting IR activity), molecular mechanisms of insulin resistance (diminished pathway signaling in obesity, diabetes), and the impact of pathway abnormalities on diabetic complications (e.g., nephropathy, neuropathy).
Core key proteins: Insulin, insulin receptor (IR), IRS1/2 (insulin receptor substrates), PI3K (phosphatidylinositol 3-kinase), Akt (protein kinase B), GLUT4 (glucose transporter), GSK3β (glycogen synthase kinase 3β), mTOR (mammalian target of rapamycin), S6K1 (ribosomal protein S6 kinase), MAPK (ERK, mitogen-activated protein kinase), PTEN (phosphatase, negative regulator), SOCS3 (suppressor of cytokine signaling 3, negative regulator), PEPCK/G6Pase (key gluconeogenic enzymes), GS (glycogen synthase).
Core key proteins: Insulin, insulin receptor (IR), IRS1/2 (insulin receptor substrates), PI3K (phosphatidylinositol 3-kinase), Akt (protein kinase B), GLUT4 (glucose transporter), GSK3β (glycogen synthase kinase 3β), mTOR (mammalian target of rapamycin), S6K1 (ribosomal protein S6 kinase), MAPK (ERK, mitogen-activated protein kinase), PTEN (phosphatase, negative regulator), SOCS3 (suppressor of cytokine signaling 3, negative regulator), PEPCK/G6Pase (key gluconeogenic enzymes), GS (glycogen synthase).
Product list
-
{{item.title}}{{item.react}}{{item.applicat}}
Product list
Product name
Reactivity
Application
Related Resource Links
Related Promotional Journal Downloads

Explore Our Recommended Popular Products
More products
30,000+ high- quality products available online
Primary Antibodies, Secondary Antibodies, mIHC Kits, ELISA Kits, Proteins, Molecular Biology Products,Cell Lines,Reagents ...
Contact Us