Core Validation Strategies
——Integrating the Antibody Validation System Proposed by the International Working Group on Antibody Validation (IWGAV) and Antibody Application Suitability Validation
Composed of international scientists in the field of protein biology, the International Working Group on Antibody Validation (IWGAV) published its first antibody validation strategy proposal titled "A Proposal for Validation of Antibodies" in Nature Methods in 2016. It put forward five "pillar validation strategies" focusing on antibody specificity, functionality, and reproducibility, providing core guidance for antibody validation in specific application scenarios.
The selection of antibody validation strategies must be based on the background data of the target, with comprehensive consideration of key factors including the target's expression abundance, spatiotemporal expression specificity, the detection threshold of the antibody itself, and applicable detection technologies. Based on these factors, the most suitable validation scheme for experimental needs is selected. Currently, the aforementioned "five core validation strategies" have been clearly marked on the official website for reference.
I. Five Core Validation Strategies (Pillar Strategies)
1. Knockout/Knockdown Strategy (KO/KD)
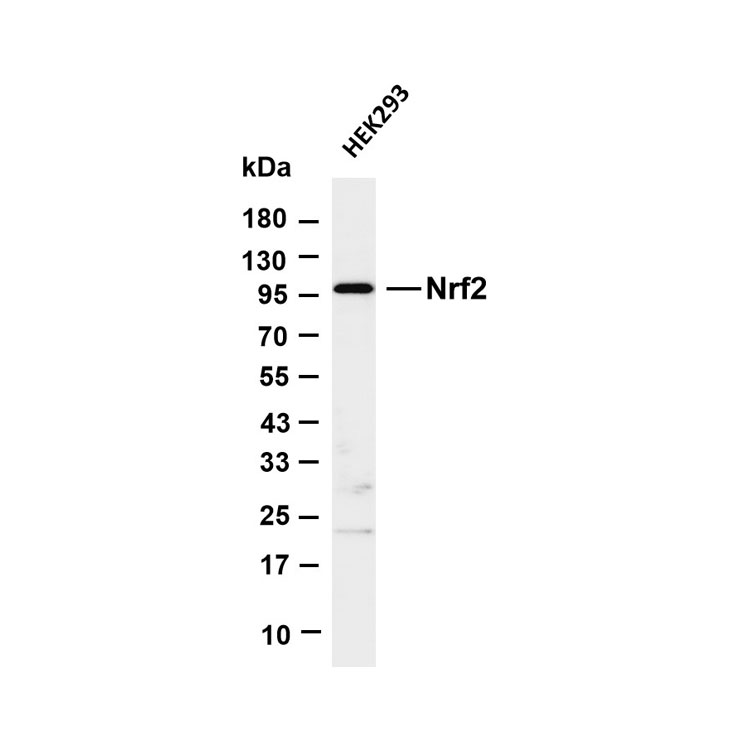
Principle: Use CRISPR/Cas9 technology to knock out the target gene, or siRNA (short-term silencing) and shRNA (long-term silencing) to knock down target gene expression, constructing KO/KD samples/cell lines that do not express the target protein as controls.
Validation Standard: In Western Blot (WB) detection, if KO/KD samples show negative signals while normal samples show positive signals, the antibody specificity is fully validated. It is a gene-level validation method with extremely high credibility and acceptability.
2. Biological Characteristic & Orthogonal Strategy
Principle: Based on biological characteristics such as natural differential protein expression, changes in gene/protein/metabolic levels, and signal pathway alterations, non-antibody-dependent methods (e.g., RNA expression detection) are used to quantify a large number of samples and analyze the correlation with antibody detection results.
Core Logic: Regulate antibody signals through changes in biological characteristics, and validate specificity through cross-method consistency.
3. Independent Antibody Validation Strategy (Comparable)
Principle: Adopt two or more independent antibodies that can recognize different epitopes of the target protein, and confirm antibody specificity by comparing or quantitatively analyzing the consistency of results.
4. Target Protein Overexpression Strategy
Principle: Modify the endogenous target gene by inserting affinity tag or fluorescent protein sequences, and conduct correlation analysis between the signal of the labeled protein and the antibody detection signal to validate antibody specificity.
5. Immunocapture/Mass Spectrometry Strategy (IP/MS)
Principle: Isolate the target protein with antibodies via Immunoprecipitation (IP) technology, and combine with Mass Spectrometry (MS) analysis to identify the target protein directly interacting with the antibody and related complex proteins.
II. Antibody Application Suitability Validation
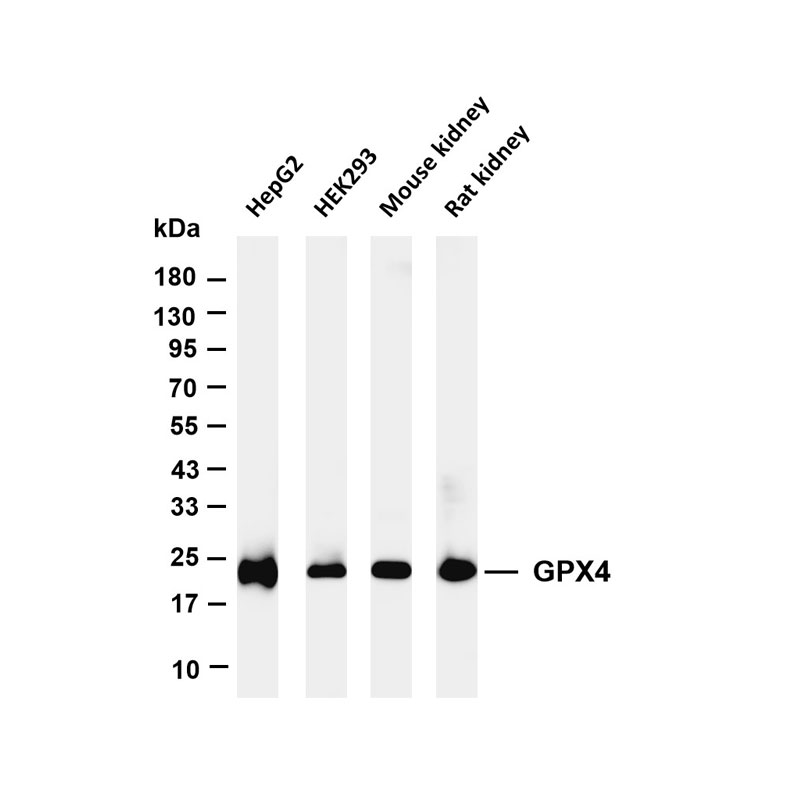
Species Cross-Reactivity Validation: Design immunogens according to the target detection species, and determine the antibody's cross-reactivity among different species through detection of samples from various species.
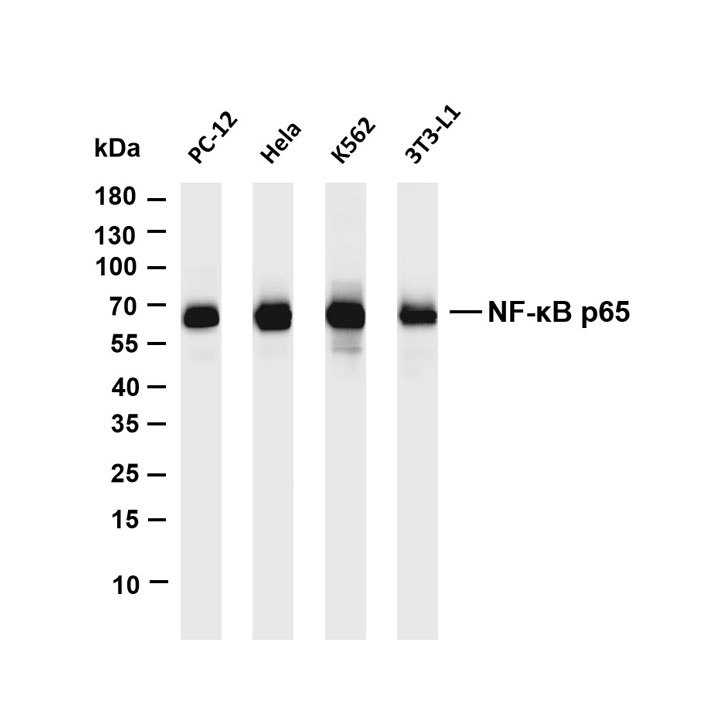
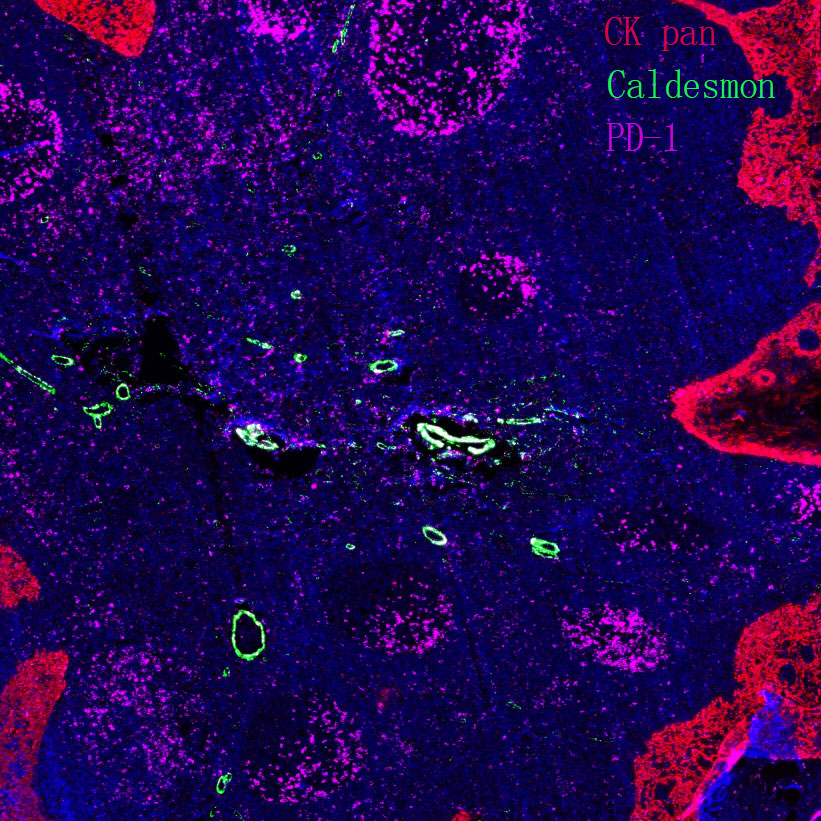
Experimental Application Scope Validation: Determine the specific experimental application requirements that the antibody can meet through detection with multiple experimental methods.
Related Promotional Journal Downloads

30,000+ high- quality products available online
Primary Antibodies, Secondary Antibodies, mIHC Kits, ELISA Kits, Proteins, Molecular Biology Products,Cell Lines,Reagents ...
Contact Us